Translational Signal Intelligence (TSI)
Signal-First Intelligence for Drug Development
>100× computational efficiency Demonstrated in quantum-enhanced optimization for patient stratification, enabling scalable covariate balancing in realistic clinical trial cohorts.
Up to 5× treatment-effect significance Observed through improved covariate balance and reduced treatment-effect dilution in post-hoc analyses of completed Phase III oncology trials.
~10× feature dimensionality reduction From thousands of genomic and transcriptomic features to compact, biologically meaningful biomarker panels without loss of predictive performance.
Near-optimal covariate balance Achieved across numerical and categorical variables, consistently outperforming randomization and classical heuristic baselines.
Why Signal Quality Matters More Than Data Volume
Strengthening machine learning by addressing biological complexity upstream
The Emerging Bottleneck
Drug development is entering a phase where data availability is no longer the primary constraint. Multimodal datasets are now routine. Yet decision confidence has not scaled accordingly. The limiting factor has shifted to signal quality:
Biologically relevant signals are sparse and non-linear
Feature spaces are combinatorial and highly redundant
Classical methods struggle to distinguish causal structure from correlation
Increasing data volume often amplifies noise rather than insight
The central challenge is no longer how much data we collect, but how effectively we extract compact, interpretable, and decision-relevant biological signals from complex systems.
Our Solution
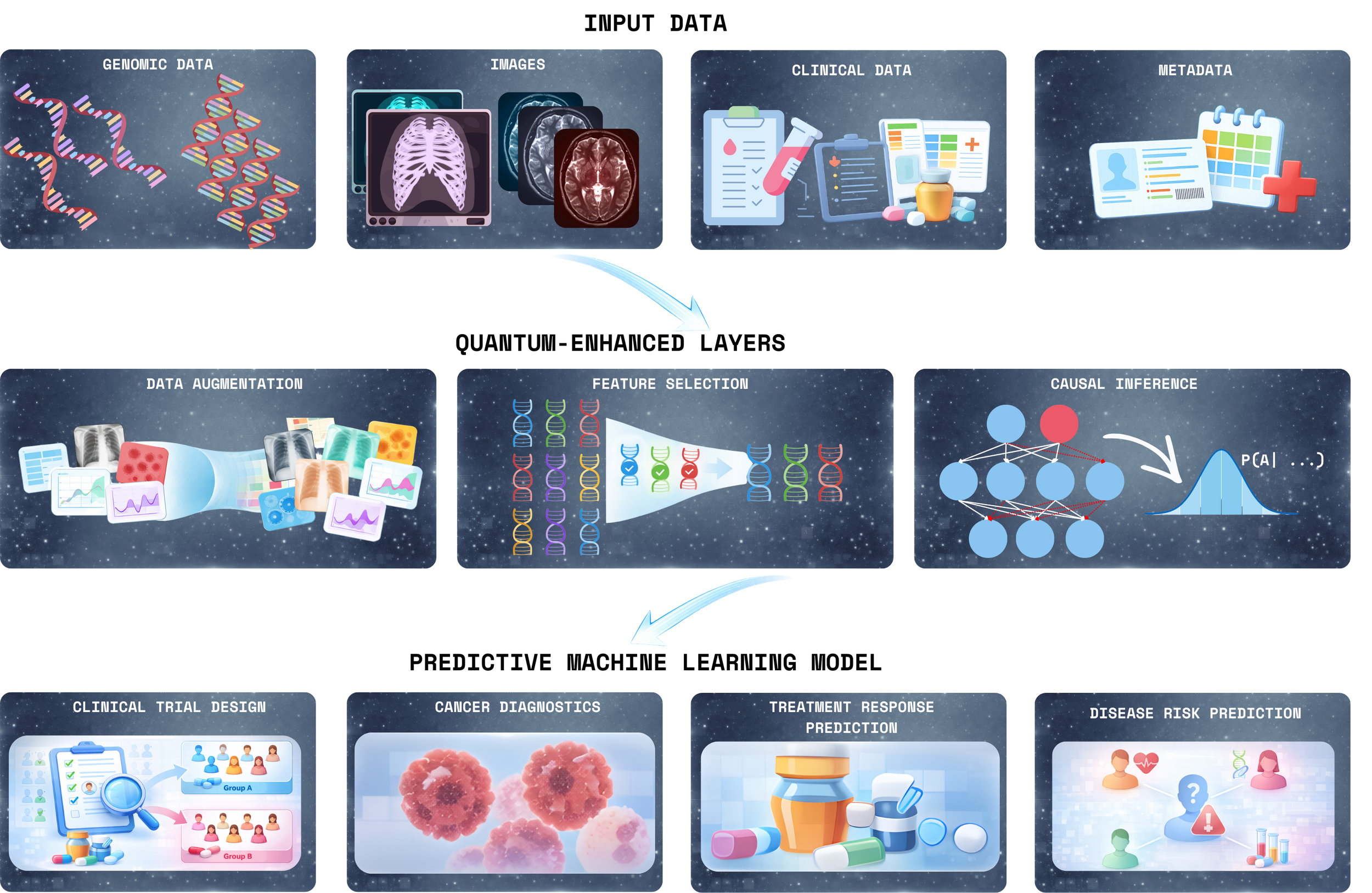
TSI is an emerging quantum-enhanced signal intelligence layer designed to operate at this bottleneck. Rather than replacing existing machine-learning pipelines, TSI strengthens them by focusing on upstream signal extraction, using:
Quantum-enhanced feature selection to identify non-redundant, high-signal biomarkers
Data augmentation techniques to stabilize learning in sparse or imbalanced datasets
Probabilistic and causal modeling to preserve interpretability and biological coherence
The result is a refined representation of biological systems that downstream classical models can use more efficiently and reliably.
Research Use Cases and Observed Impact
Applied quantum research in biomedical systems
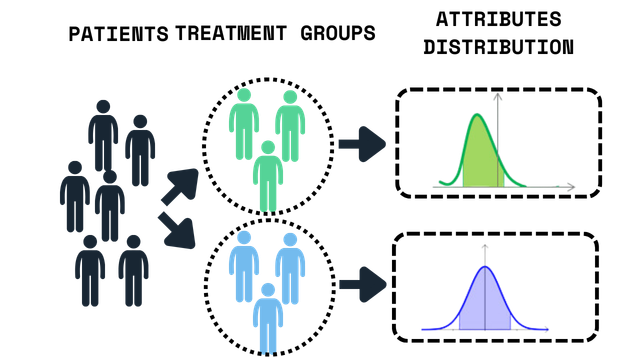
Clinical Trial Design & Patient Stratification
Optimization-based patient stratification under biological heterogeneity.
What was Demonstrated
Hybrid quantum–classical optimization minimized covariate imbalance
Near-optimal balance at scales where classical solvers fail
No changes to endpoints or analysis plans
Observed Impact
Reduced treatment-effect dilution
Up to fivefold improvement in statistical significance
Earlier, more reliable go/no-go decisions
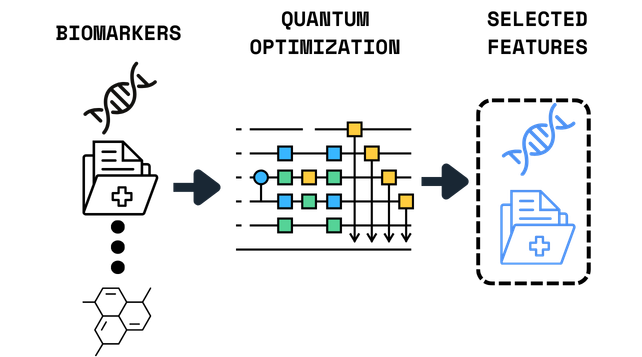
Biomarker Discovery in High-Dimensional Data
Feature selection across thousands of genomic and transcriptomic variables.
What was Demonstrated
>5,000 features reduced to ~15 biomarkers
Biologically coherent and interpretable panels
Improved or preserved downstream model performance
Observed Impact
Experimentally tractable biomarker panels
Improved translational confidence
Foundation for diagnostic
Earlier, more reliable go/no-go decisions
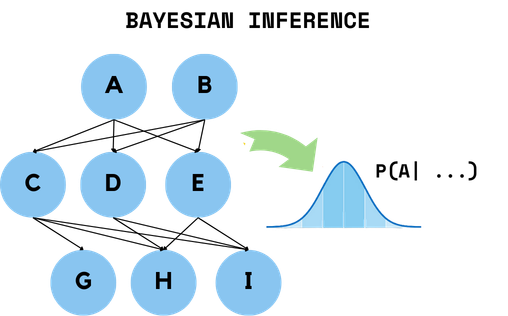
Bayesian Modelling of Disease Dynamics
Probabilistic modelling of disease evolution and symptom prediction.
What was Demonstrated
Squared amplification of rare events
Efficient scaling of network structure learning
Observed Impact
Interpretable and scalable learning of probabilistic models linking symptom evolution
Relevance for patient monitoring and treatment selection
NIH CHALLENGE WINNERS
This work was recognized as a winning project in the NIH Quantum Computing Challenge, a national initiative evaluating practical, high-impact applications of quantum and quantum-inspired methods in biomedical research.